How to Read Gel Electrophoresis Cut Vs Uncut
Congratulations, you have a plasmid expressing your cistron of involvement (YGOI) and are ready to swoop into your functional experiments! Whether you've cloned the plasmid yourself or obtained it from a colleague down the hall, information technology is e'er a good idea to take some time to ostend that you are working with the correct construct, and verify that the plasmid yous received matches the expected sequence. Hither at Addgene, we use NGS-based quality control to ostend the sequence of all the plasmids we distribute. This method is fourth dimension-intensive, so nosotros recommend a multifariousness of means to screen and verify your plasmids. Hither, we'll embrace restriction digest analysis. Diagnostic digests tin can exist used to ostend the rough structure of the plasmid based on the predicted sizes and organization of different featureswithin the plasmid. Restriction analysis can also be used successfully even if you don't have the full plasmid sequence. Once you have purified plasmid DNA, this method can be washed correct in your lab in less than a solar day. Diagnostic restriction digests are comprised of 2 separate steps: 1) incubating your DNA with brake enzymes which cleave the DNA molecules at specific sites and 2) running the reaction on an agarose gel to determine the relative sizes of the resulting Dna fragments. Restriction digests are ordinarily used to confirm the presence of an insert in a detail vector by excising the insert from the backbone. To practise this, you'll use enzymes with brake sites that flank the insert. You lot volition need to know both the estimate size of the vector backbone besides equally the predicted size of the insert. You tin can search NCBI for YGOI to observe the particular reference sequence if necessary. Sentinel this video for a quick overview of how to clarify a restriction digest: The following tips volition make information technology easier for you lot to obtain a useful and informative diagnostic restriction assimilate. More than Plasmid Eductional Resources: 
Diagnostic restriction digest
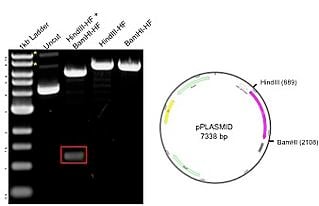 The example plasmid on the right has a total size of 7.3kb, including a 1.2 kb insert. The plasmid was digested with ii unique enzymes (HindIII and BamHI) and run on an agarose gel. The resulting gel image includes a 1kb ladder (lane one) that has bands ranging from nearly 500bp to 10kb, with the 3.0kb fragment having increased intensity to serve equally a reference band. The uncut Deoxyribonucleic acid (lane 2) shows three possible plasmid conformations, with relaxed and nicked marked with asterisks (*). When the plasmid is digested with eitherHindIII and BamHI alone (lanes 4-5), at that place is a single band of 7.3 kb representing the full size of the plasmid. The double digest with both HindIII and BamHI (lane 3) produces bands at 6kb and 1.2kb (red box), matching the backbone and insert, respectively. The results on the gel correspond to the predicted sizes.
The example plasmid on the right has a total size of 7.3kb, including a 1.2 kb insert. The plasmid was digested with ii unique enzymes (HindIII and BamHI) and run on an agarose gel. The resulting gel image includes a 1kb ladder (lane one) that has bands ranging from nearly 500bp to 10kb, with the 3.0kb fragment having increased intensity to serve equally a reference band. The uncut Deoxyribonucleic acid (lane 2) shows three possible plasmid conformations, with relaxed and nicked marked with asterisks (*). When the plasmid is digested with eitherHindIII and BamHI alone (lanes 4-5), at that place is a single band of 7.3 kb representing the full size of the plasmid. The double digest with both HindIII and BamHI (lane 3) produces bands at 6kb and 1.2kb (red box), matching the backbone and insert, respectively. The results on the gel correspond to the predicted sizes.Restriction digest tips and tricks:
For your digest:
For your gel:

Source: https://blog.addgene.org/plasmids-101-how-to-verify-your-plasmid
0 Response to "How to Read Gel Electrophoresis Cut Vs Uncut"
Post a Comment